-
PDF
- Split View
-
Views
-
Cite
Cite
Chii Shyang Fong, Mark D. Temple, Nazif Alic, Joyce Chiu, Moritz Durchdewald, Geoffrey W. Thorpe, Vincent J. Higgins, Ian W. Dawes, Oxidant-induced cell-cycle delay in Saccharomyces cerevisiae: the involvement of the SWI6 transcription factor, FEMS Yeast Research, Volume 8, Issue 3, May 2008, Pages 386–399, https://doi.org/10.1111/j.1567-1364.2007.00349.x
Close - Share Icon Share
Abstract
Cells treated with low doses of linoleic acid hydroperoxide (LoaOOH) exhibit a cell-cycle delay that may provide a mechanism to overcome oxidative stress. Strains sensitive to LoaOOH from the genome-wide deletion collection were screened to identify deletants in which the cell-cycle delay phenotype was reduced. Forty-seven deletants were identified that were unable to mount the normal delay response, implicating the product of the deleted gene in the oxidant-mediated cell-cycle delay of the wild-type. Of these genes, SWI6 was of particular interest due to its role in cell-cycle progression through Start. The swi6 deletant strain was delayed on entry into the cell cycle in the absence of an oxidant, and oxidant addition caused no further delay. Transforming the swi6 deletant with SWI6 on a plasmid restored the G1 arrest in response to LoaOOH, indicating that Swi6p is involved in oxidant sensing leading to cell division delay. Micro-array studies identified genes whose expression in response to LoaOOH depended on SWI6. The screening identified 77 genes that were upregulated in the wild-type strain and concurrently downregulated in the swi6 deletant treated with LoaOOH. These data show that functions such as heat shock response, and glucose transport are involved in the response.
Introduction
Cells exhibit a range of responses to oxidative stress, including adaptation to increased resistance (Collinson & Dawes, 1992), cell-cycle progression delay (Flattery-O'Brien & Dawes, 1998) and widespread changes in gene expression (Gasch et al., 2000) while at higher concentrations they are no longer able to survive and undergo a form of apoptosis (Ludovico et al., 2005). They also show similar responses to lipid peroxides formed as a result of reactive oxygen species (ROS) damaging unsaturated lipids. Here, the authors report on genes involved in the response of Saccharomyces cerevisiae to linoleic acid hydroperoxide (13-hydroperoxylinoleic acid; LoaOOH), which is an oxidant that is an oxidative breakdown product of lipid peroxidation, and one of the most toxic peroxides to yeast cells (Evans et al., 1998; Aoshima et al., 1999). LoaOOH is a product of free radical attack on a long-chain unsaturated fatty acid, and it has maximal toxicity to cells undergoing respiration. Like other lipid peroxides, it is reactive and, in the cell, breaks down into other highly reactive products including epoxides and reactive aldehydes such as malondialdehyde and 4-hydroxynonenal (Evans et al., 1998; Dickinson & Forman, 2002).
LoaOOH treatment of wild-type cells leads to a G1 delay in cell-cycle progression before the Start checkpoint. During this delay cells may mount antioxidant defences and systems to elicit adequate repair of oxidant damage before re-engaging in the cell cycle (Shackelford et al., 2000). This leads to accumulation of unbudded cells in asynchronous populations, together with a budding and replication delay in synchronous ones (Mendenhall & Hodge, 1998; Alic et al., 2001). Currently, little is known about the genes or processes that are involved in this cell-cycle delay. Only two genes were previously implicated in the G1 delay in response to LoaOOH or other oxidants. These were OCA1 and PEX17 because strains deleted for these genes did not show cell-cycle delay in response to LoaOOH (Alic et al., 2001).
For cells to pass Start and commit to cell-cycle progression, the two transcription factor complexes SBF (Swi4p/Swi6p-dependent cell-cycle box-binding factor) and MBF (Mbp1p/Swi6p-dependent cell-cycle box-binding factor) are required. These form part of an underlying transcriptional regulatory network of nine transcription factors that control the expression of the cyclins and oscillations of the cyclin/CDK activities during the cell cycle (Simon et al., 2001). The activation of Swi4p is dependent on Cln3p/Cdc28p activity as cells reach a critical size (Koch et al., 1996) and Swi4p is unable to bind to promoters in the absence of Swi6p (Baetz & Andrews, 1999). Together, SBF and MBF control the activation of many G1/S phase specific-genes. The cyclin genes Cln1p and Cln2p that associate with the major cyclin-dependent kinase Cdc28p are targets of SBF/MBF and SBF, respectively. Predominantly, the relevant genes involved in cell-cycle control are activated by either SBF or MBF, whereas genes involved in cell-wall biogenesis, budding, cytokinesis, histones, chromatin modifiers and telomere length are activated by SBF alone, and those involved in DNA replication are activated by MBF (Simon et al., 2001).
Interestingly, a systematic screen of the genome-wide set of deletion mutants each with a deletion in one nonessential gene (Winzeler et al., 1999) revealed that 256 deletants were sensitive to LoaOOH (Thorpe et al., 2004). These studies showed that lipid and carbohydrate metabolism is crucial for LoaOOH tolerance, the peroxisome may be the site of LoaOOH detoxification and that energy from the tricarboxylic acid cycle is required to maintain active defenses against lipid peroxidation (Thorpe et al., 2004). Interestingly, they also showed that the swi6 deletion strain was sensitive to LoaOOH.
While the involvement of SWI6 indicates that one or both of the canonical transcription factor complexes SBF and MBF may be involved in maintaining resistance to LoaOOH, it is not known whether it is involved in the G1 checkpoint arrest and what other systems may be involved. Because cell-division delay is a phenotype that is not easy to determine in a high-throughput screening, a systematic evaluation has been carried out of LoaOOH-induced cell division arrest in deletants of S. cerevisiae that are sensitive to LoaOOH treatment because such mutants are most likely to be the ones affected in one or more of the cellular responses to the ROS.
Materials and methods
Strain and plasmids
The S. cerevisiae deletants used in this study were in the homozygous diploid strain BY4743 genetic background (MATa/MATα; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; met15Δ0/MET15; LYS2/lys2Δ0; ura3Δ0/ura3Δ0) (Brachmann et al., 1998). These deletants were provided by the European S. cerevisiae Archive of Functional Analysis (EUROSCARF). For microarray analysis, the wild-type S. cerevisiae haploid strain BY4741 (MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0; Euroscarf) (Brachmann et al., 1998) was used, together with the swi6 deletant strain (MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 swi6∷kanMX4).
SWI6 coding sequence and its flanking sequences (1 kb upstream and 0.5 kb downstream) were amplified by PCR and cloned into the pRS415 CEN LEU2 vector. The resulting plasmid pSWI6 and the vector control pRS415 were transformed into the swi6 deletant and LEU2+ transformants were selected. The vector control pRS415 was also transformed into swi6 deletant strain as a control.
Media and chemicals
YEPD medium contained 2% (w/v) glucose, 2% (w/v) bactopeptone and 1% yeast extract. YEPG medium contained 2% (w/v) bactopeptone, 1% yeast extract and 3% (v/v) glycerol. Synthetic complete (SC) medium contained 2% (w/v) glucose, 0.17% yeast nitrogen base, 0.5% ammonium sulphate, 0.074% complete supplement mixture without tryptophan (CSM-TRP) and 0.01% tryptophan; 2% (w/v) agar was added to solidify the media. Cultures were shaken at 300 r.p.m. and incubated at 30 °C, unless otherwise noted. Linoleic acid hydroperoxide (LoaOOH) stock was prepared and assayed as described by Evans et al., (1998) and stored at −20 °C in methanol. Synchronization of MATa strains using α-factor, oxidant treatment and release, and determination of α-factor-resistant cells was performed as described previously (Alic et al., 2001).
Screening of mutants
SC medium (100 mL) was inoculated with 10 μL of an overnight YEPD starter culture and incubated at 30 °C to an OD600 nm of 0.2. Cultures were equally divided into two flasks and incubated until the OD600 nm reached 0.3–0.4. LoaOOH (to 0.01 mM) was added to one of the flasks, leaving the other as an untreated control. Samples were collected 60 min after treatment for budding index (BI) estimation. Cell culture (1 mL) was collected and cells were fixed in 3 mL of ice-chilled ethanol. Cells were then washed in 1 mL of 0.2 M Tris-HCl (pH7.5), resuspended in 20 μL of 0.2 M Tris-HCl (pH7.5) and maintained at 4 °C until counting. Where clumping occurred, washed cells were sonicated with six pulses of 5 s each at 30% power in a M250 Branson digital sonifier (Branson) before resuspension. Before counting, 5 μL of fixed, resuspended cells were mixed with 4′, 6-diamidino-2-phenylindole, dihydrochloride (DAPI) and were viewed under phase contrast in a fluorescence microscope (Olympus BX60) using a 330–385 nm filter to detect DAPI fluorescence. At least 400 cells were counted for each sample.
Cell-cycle analysis
Strains harbouring plasmids pRS415 or pSWI6 were cultured in SC-leu medium at 30 °C until the OD600 nm reached 0.2. α-Factor was added to 1 μg mL−1 and cultures were synchronized by further incubation at 30 °C for 2 h. Cells were subsequently washed and resuspended in phosphate-buffered saline (PBS; 8 g L−1 NaCl, 0.2 g L−1 KCl, 1.44 g L−1 Na2HPO4, 0.24 g L−1 KH2PO4 adjusted to pH 7.4 with HCl). The cultures were divided into equal aliquots and LoaOOH was added to a final concentration of 0.02 mM in one, while an equal volume of methanol was added to the other culture. Cultures were incubated with shaking at 30 °C for 30 min and were then washed once with PBS. Cells were resuspended in 20 mL SC-leu medium and 1 mL samples were taken at 15-min intervals for 135 min. Cell samples were spun down and fixed with 1 mL of ice-cold 70% (v/v) ethanol and resuspended in 1 mL of 50 mM Tris-HCl, pH 7.5. Cells were subsequently sonicated in an M250 Branson digital sonifier (Branson) for 20 s at 30% amplitude to disperse clumps, washed once with 1 mL of 50 mM Tris-HCl pH 7.5 and resuspended in 20 μL of the same buffer. Five microliters of cells were mixed with 1 μL of 100 μg mL−1 DAPI and were observed under × 100 phase contrast using a BX60 Olympus microscope (Olympus, Japan).
Microarray analysis
Cultures were grown overnight to an OD600 nm of 0.2, divided into 200 mL aliquots into prewarmed flasks and allowed to grow to an OD600 nm of 0.4. The aliquots were then treated with 0.03 mM LoaOOH for 60 min because this was found to be the optimal treatment to measure the responsive phenotype previously (Alic et al., 2003). The reference RNA pool was obtained from five separate 200 mL aliquots of untreated cells harvested at time zero. The control culture was treated with methanol only, equivalent to the volume required for LoaOOH treatment. In comparing the transcriptional response of the swi6 deletant with the wild type, biological duplicates were obtained for each strain. Samples (200 mL) were harvested by mixing with 40 g of −80 °C ice in prechilled tubes and centrifuged (5 min at 3345 g) at 4 °C. The cells were then rapidly frozen in an ethanol bath at −80 °C. The RNA was isolated by breaking cells in Trizol reagent (Gibco BRL, Life Technologies, MD) in a mini-bead beater in the presence of acid-washed glass beads at 4 °C, and extracted according to the manufacturer's instructions. The RNA was further purified using the RNeasy kit (Qiagen, Dusseldorf, Germany), including DNAse I treatment. RNA quality was checked by agarose gel electrophoresis and OD260 nm/280 nm determination, after which the reference samples were pooled.
Spotted oligonucleotide microarrays were obtained from the Clive and Vera Ramaciotti Centre for Gene Function Analysis (Sydney, Australia). Oligonucleotide probes (MWG Biotech, Ebersberg, Germany) addressing 6250 yeast ORFs were printed in duplicate on epoxy-coated glass substrates (Eppendorf, Hamburg, Germany) and were blocked immediately before use with ethanediol according to the manufacturer's instructions. Each comparison was performed in duplicate.
Labelling and hybridization were carried out according to a modification of the protocol described by Hughes et al., (2001). Twenty grams of total RNA was reverse-transcribed incorporating 5-(3-aminoallyl)-dUTP (Sigma). The cDNA was labelled by coupling the aminoallyl-dUTP to N-hydroxy succinimide esters of either Cy3 or Cy5 (Amersham Biosciences). The labelled cDNA of the samples to be compared was mixed and hybridized to the array overnight at 37 °C in DIG Easy Hyb (Roche Applied Science) containing 0.5 mg mL−1Escherichia coli tRNA and 0.5 mg mL−1 denatured herring sperm DNA. The slides were then washed three times for 20 min in SSC (3 M sodium chloride, 0.3 M trisodium citrate, pH 7.5) containing 0.1% sodium dodecyl sulphate at 50 °C, rinsed several times in SSC and dried by centrifugation. The slides were scanned using an ArrayWoRx E Biochip Reader (Applied Precision, WA) and technical duplicates were performed including a dye-swap.
Data processing
Image analysis was performed in genepix pro 3.0 (Axon Instruments). The signal for a gene was deemed ‘present’ if no artefacts were associated with the spot, and the program could identify the spot intensity above the background intensity. Data were imported into genespring 5.0 (Silicon Genetics), where all further analysis was performed. Data were normalized by the LOWESS normalization method. Only the genes ‘present’ on all six slides were considered further. To identify genes with twofold different expression, only the genes ‘present’ on both duplicates of a given condition were considered. Functional enrichment in the sets obtained was determined using funspec (Robinson et al., 2002) and all available databases. For information on individual gene products, the SGD database was consulted. Gene lists derived from the microarray data are given in supplementary Table S2.
Results
Genes involved in cell-cycle delay following LoaOOH treatment
Delay of cell-cycle progression is thought to allow cells time to mount defence and repair systems (Hartwell & Weinert, 1989; Shackelford et al., 2000); hence, it was reasoned that the inability of cells to delay would be likely to contribute to LoaOOH sensitivity. Because budding is an inherent property of S. cerevisiae that occurs only after cells have passed through Start and after initiation of cell division (Zettel et al., 2003), a systematic evaluation of the BI of 111 of the most sensitive deletants (Thorpe et al., 2004) in response to a sublethal dose of LoaOOH was carried out to assess their cell-cycle response phenotype.
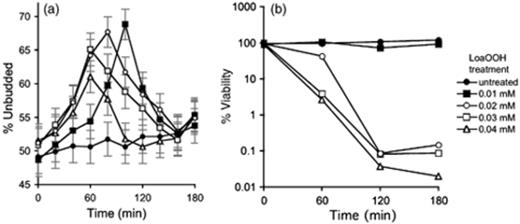
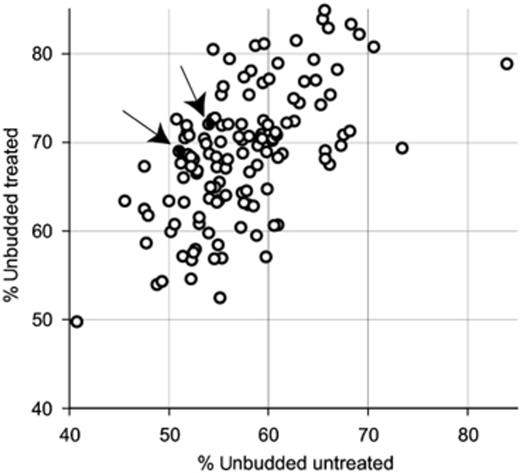
Exponentially growing cells of each sensitive mutant were treated with a range of concentrations of LoaOOH (0–0.04 mM) and samples were harvested for estimation of BI at 30-min intervals over a 3-h period. Harvested cells were fixed in ethanol, stained with DAPI and the BI of each mutant was determined using fluorescence microscopy. Representative images used for these analyses are shown in Fig. 1. The BI (BI=proportion of cells that are budded) was calculated for the wild type and each deletant strain. Untreated wild-type cells had a roughly equal distribution of budded and unbudded cells (BI=0.52), whereas wild-type cells treated with 0.01 mM LoaOOH exhibited an accumulation of 69% unbudded cells (BI=0.69) as shown in Fig. 2a. At this dose, the viability of LoaOOH-treated cells remained high during the treatment as shown in Fig. remained high during the treatment as shown in Fig. 2b. From these data, it is clear that cells treated with 0.01 mM LoaOOH exhibited both high accumulation (>70%) of G1 unbudded cells at c. 60 min and high cell viability relative to the untreated sample. The increase in unbudded cells is therefore an indicator of cell-cycle delay that is caused by treatment with LoaOOH, and hence the percentage increase in unbudded cells on LoaOOH (ΔUBLoaOOH) was determined for all strains. The data obtained from the mutant screen are shown in Fig. 3 to illustrate the distribution of budding percentages obtained for the mutant strains. From this it can be seen that some mutants have a substantially greater percentage of unbudded cells relative to the wild-type strain without any treatment, indicating that the mutations may be causing some delay in cell-cycle progression through Start into the S phase. What is of greater interest here is whether any of these mutants showed a significant change in the percentage of unbudded cells after treatment with LoaOOH.
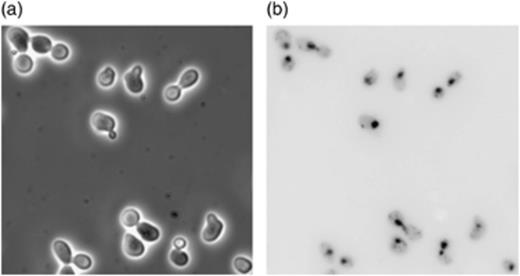
Microscopic analysis of BY4743 swi6 deletant cell budding. (a) Phase-contrast microscopy image of cells. (b) Cells stained with DAPI and viewed under fluorescence. Cells were scored as budded cells if, under phase-contrast microscopy, both the mother and the bud were visible and under fluorescence microscopy only the mother cell but not the bud exhibited DAPI staining of the nucleus.
Response of wild-type cells to LoaOOH treatment. (a) The percentage budding of exponential phase wild-type cells treated with various concentrations of LoaOOH for 180 min. Harvested cells were fixed, stained with DAPI and buds were counted for each treatment. Data are means and SD of duplicate experiments. (b) A representative experiment showing the viability of wild-type cells. Samples were diluted and plated onto YEPD plates to monitor cell viability.
Graph of the percentage of unbudded cells for LoaOOH-stressed (ordinate) and unstressed (abscissa) conditions. Individual deletion strains are represented by open circles, and representative wild-type ones by closed circles indicated by arrows.
The average ΔUBLoaOOH of the wild type (seven replicates) was 17.5% (SD 4.75). This large increase in the percentage of unbudded cells reflects the extent of delayed passage through Start in response to LoaOOH treatment. A deletant strain was considered to be affected in this cell-cycle delay if the measured ΔUBLoaOOH was at least two SDs (9.5%) lower than the wild type response, i.e. all deletants that exhibited a ΔUBLoaOOH of <8. Table 1 lists the 47 deletants that exhibited a significantly lower ΔUBLoaOOH than the wild type, together with the response of the wild-type for comparison. It also lists the sensitivity of the individual deletants to LoaOOH as determined by Thorpe et al., (2004).
Deletant strains of Saccharomyces cerevisiae that exhibited an absence of G1 checkpoint in response to LoaOOH treatment
| ORF | Gene | ΔUBLoaOOH | ΔBIuntreated | LoaOOH sensitivity |
| BY4743 | Wild-type | 17.5 | 0 | Not sensitive |
| YMR169C | ALD3 | 2.79 | −3.27 | 4 |
| YJL115W | ASF1 | 5.05 | 6.51 | 4 |
| YER177W | BMH1 | 5.88 | 4.19 | 4 |
| YER141W | COX15 | 0.38 | 2.11 | 6 |
| YGR036C | CWH8 | 0.31 | 10.13 | 6 |
| YIL065C | FIS1 | 2.29 | −4.2 | 6 |
| YOL051W | GAL11 | −4.44 | 4.24 | 3 |
| YGR163W | GTR2 | 0.09 | 5.65 | 6 |
| YLR192C | HCR1 | 2.33 | 8.3 | 7 |
| YDR174W | HMO1 | 4.36 | 8.33 | 3 |
| YDL115C | IWR1 | 6.56 | 10.96 | 4 |
| YLR244C | MAP1 | 7.72 | 4.46 | 2 |
| YLR320W | MMS22 | 5.51 | −15.91 | 5 |
| YKL009W | MRT4 | 4.15 | 14.49 | 6 |
| YNL099C | OCA1 | 1.18 | 2.36 | 7 |
| YJR073C | OPI3 | 3.02 | 3.93 | 3 |
| YBR035C | PDX3 | 2.1 | 13.3 | 2 |
| YLR148W | PEP3 | 0.82 | 4.29 | 6 |
| YDL065C | PEX19 | 0.32 | −1.25 | 6 |
| YIL107C | PFK26 | 3.43 | −2.1 | 2 |
| YGL025C | PGD1 | −1.74 | −0.13 | 4 |
| YDL006W | PTC1 | 3.36 | 5.89 | 6 |
| YJL121C | RPE1 | 7.8 | −1.36 | 2 |
| YLL002W | RTT109 | −2.46 | −9.45 | 6 |
| YNL032W | SIW14 | 4.33 | 1.63 | 7 |
| YBR289W | SNF5 | 3.36 | 8.1 | 1 |
| YHL025W | SNF6 | 6.49 | 0.93 | 3 |
| YGR104C | SRB5 | −1.85 | 6.92 | 4 |
| YPL057C | SUR1 | 2.66 | 9.44 | 6 |
| YJL176C | SWI3 | 1.33 | 10.63 | 1 |
| YLR182W | SWI6 | 3.08 | 13.75 | 7 |
| YBR069C | TAT1 | 1.52 | 21.51 | 2 |
| YLR237W | THI7 | 2.33 | 5.97 | 7 |
| YCR053W | THR4 | 6.09 | −2.06 | 3 |
| YPR163C | TIF3 | 4.1 | 9.91 | 6 |
| YPR074C | TKL1 | 6.78 | 5.9 | 1 |
| YOR332W | VMA4 | 4.63 | 4.06 | 4 |
| YML007W | YAP1 | 4.11 | 7.3 | 3 |
| YBR285W | YBR285W | 2.33 | 1.63 | 7 |
| YCR061W | YCR061W | 2 | 2.3 | 7 |
| YCR095C | YCR095C | 2 | 3.63 | 7 |
| YDL173W | YDL173W | 5.34 | 1.18 | 3 |
| YIL077C | YIL077C | −6.08 | −0.8 | 6 |
| YLR456W | YLR456W | 4.12 | 1.95 | 4 |
| YNL080C | YNL080C | 1.38 | −3.54 | 5 |
| YOR135C | YOR135C | 6.58 | 1.12 | 3 |
| Overlap | YGR064W | 4.61 | 2.34 | 4 |
| SPT4 |
| ORF | Gene | ΔUBLoaOOH | ΔBIuntreated | LoaOOH sensitivity |
| BY4743 | Wild-type | 17.5 | 0 | Not sensitive |
| YMR169C | ALD3 | 2.79 | −3.27 | 4 |
| YJL115W | ASF1 | 5.05 | 6.51 | 4 |
| YER177W | BMH1 | 5.88 | 4.19 | 4 |
| YER141W | COX15 | 0.38 | 2.11 | 6 |
| YGR036C | CWH8 | 0.31 | 10.13 | 6 |
| YIL065C | FIS1 | 2.29 | −4.2 | 6 |
| YOL051W | GAL11 | −4.44 | 4.24 | 3 |
| YGR163W | GTR2 | 0.09 | 5.65 | 6 |
| YLR192C | HCR1 | 2.33 | 8.3 | 7 |
| YDR174W | HMO1 | 4.36 | 8.33 | 3 |
| YDL115C | IWR1 | 6.56 | 10.96 | 4 |
| YLR244C | MAP1 | 7.72 | 4.46 | 2 |
| YLR320W | MMS22 | 5.51 | −15.91 | 5 |
| YKL009W | MRT4 | 4.15 | 14.49 | 6 |
| YNL099C | OCA1 | 1.18 | 2.36 | 7 |
| YJR073C | OPI3 | 3.02 | 3.93 | 3 |
| YBR035C | PDX3 | 2.1 | 13.3 | 2 |
| YLR148W | PEP3 | 0.82 | 4.29 | 6 |
| YDL065C | PEX19 | 0.32 | −1.25 | 6 |
| YIL107C | PFK26 | 3.43 | −2.1 | 2 |
| YGL025C | PGD1 | −1.74 | −0.13 | 4 |
| YDL006W | PTC1 | 3.36 | 5.89 | 6 |
| YJL121C | RPE1 | 7.8 | −1.36 | 2 |
| YLL002W | RTT109 | −2.46 | −9.45 | 6 |
| YNL032W | SIW14 | 4.33 | 1.63 | 7 |
| YBR289W | SNF5 | 3.36 | 8.1 | 1 |
| YHL025W | SNF6 | 6.49 | 0.93 | 3 |
| YGR104C | SRB5 | −1.85 | 6.92 | 4 |
| YPL057C | SUR1 | 2.66 | 9.44 | 6 |
| YJL176C | SWI3 | 1.33 | 10.63 | 1 |
| YLR182W | SWI6 | 3.08 | 13.75 | 7 |
| YBR069C | TAT1 | 1.52 | 21.51 | 2 |
| YLR237W | THI7 | 2.33 | 5.97 | 7 |
| YCR053W | THR4 | 6.09 | −2.06 | 3 |
| YPR163C | TIF3 | 4.1 | 9.91 | 6 |
| YPR074C | TKL1 | 6.78 | 5.9 | 1 |
| YOR332W | VMA4 | 4.63 | 4.06 | 4 |
| YML007W | YAP1 | 4.11 | 7.3 | 3 |
| YBR285W | YBR285W | 2.33 | 1.63 | 7 |
| YCR061W | YCR061W | 2 | 2.3 | 7 |
| YCR095C | YCR095C | 2 | 3.63 | 7 |
| YDL173W | YDL173W | 5.34 | 1.18 | 3 |
| YIL077C | YIL077C | −6.08 | −0.8 | 6 |
| YLR456W | YLR456W | 4.12 | 1.95 | 4 |
| YNL080C | YNL080C | 1.38 | −3.54 | 5 |
| YOR135C | YOR135C | 6.58 | 1.12 | 3 |
| Overlap | YGR064W | 4.61 | 2.34 | 4 |
| SPT4 |
Data are given as sensitivity to the change in percent budding as a result of treatment with 0.01 mM LoaOOH for one hour (ΔUBLoaOOH). The difference in percentage buds of the untreated wild-type (ΔBIuntreated) is also given, together with the sensitivity of the strains to LoaOOH on a scale 1 (most sensitive) to 7 (least sensitive).
Deletant strains of Saccharomyces cerevisiae that exhibited an absence of G1 checkpoint in response to LoaOOH treatment
| ORF | Gene | ΔUBLoaOOH | ΔBIuntreated | LoaOOH sensitivity |
| BY4743 | Wild-type | 17.5 | 0 | Not sensitive |
| YMR169C | ALD3 | 2.79 | −3.27 | 4 |
| YJL115W | ASF1 | 5.05 | 6.51 | 4 |
| YER177W | BMH1 | 5.88 | 4.19 | 4 |
| YER141W | COX15 | 0.38 | 2.11 | 6 |
| YGR036C | CWH8 | 0.31 | 10.13 | 6 |
| YIL065C | FIS1 | 2.29 | −4.2 | 6 |
| YOL051W | GAL11 | −4.44 | 4.24 | 3 |
| YGR163W | GTR2 | 0.09 | 5.65 | 6 |
| YLR192C | HCR1 | 2.33 | 8.3 | 7 |
| YDR174W | HMO1 | 4.36 | 8.33 | 3 |
| YDL115C | IWR1 | 6.56 | 10.96 | 4 |
| YLR244C | MAP1 | 7.72 | 4.46 | 2 |
| YLR320W | MMS22 | 5.51 | −15.91 | 5 |
| YKL009W | MRT4 | 4.15 | 14.49 | 6 |
| YNL099C | OCA1 | 1.18 | 2.36 | 7 |
| YJR073C | OPI3 | 3.02 | 3.93 | 3 |
| YBR035C | PDX3 | 2.1 | 13.3 | 2 |
| YLR148W | PEP3 | 0.82 | 4.29 | 6 |
| YDL065C | PEX19 | 0.32 | −1.25 | 6 |
| YIL107C | PFK26 | 3.43 | −2.1 | 2 |
| YGL025C | PGD1 | −1.74 | −0.13 | 4 |
| YDL006W | PTC1 | 3.36 | 5.89 | 6 |
| YJL121C | RPE1 | 7.8 | −1.36 | 2 |
| YLL002W | RTT109 | −2.46 | −9.45 | 6 |
| YNL032W | SIW14 | 4.33 | 1.63 | 7 |
| YBR289W | SNF5 | 3.36 | 8.1 | 1 |
| YHL025W | SNF6 | 6.49 | 0.93 | 3 |
| YGR104C | SRB5 | −1.85 | 6.92 | 4 |
| YPL057C | SUR1 | 2.66 | 9.44 | 6 |
| YJL176C | SWI3 | 1.33 | 10.63 | 1 |
| YLR182W | SWI6 | 3.08 | 13.75 | 7 |
| YBR069C | TAT1 | 1.52 | 21.51 | 2 |
| YLR237W | THI7 | 2.33 | 5.97 | 7 |
| YCR053W | THR4 | 6.09 | −2.06 | 3 |
| YPR163C | TIF3 | 4.1 | 9.91 | 6 |
| YPR074C | TKL1 | 6.78 | 5.9 | 1 |
| YOR332W | VMA4 | 4.63 | 4.06 | 4 |
| YML007W | YAP1 | 4.11 | 7.3 | 3 |
| YBR285W | YBR285W | 2.33 | 1.63 | 7 |
| YCR061W | YCR061W | 2 | 2.3 | 7 |
| YCR095C | YCR095C | 2 | 3.63 | 7 |
| YDL173W | YDL173W | 5.34 | 1.18 | 3 |
| YIL077C | YIL077C | −6.08 | −0.8 | 6 |
| YLR456W | YLR456W | 4.12 | 1.95 | 4 |
| YNL080C | YNL080C | 1.38 | −3.54 | 5 |
| YOR135C | YOR135C | 6.58 | 1.12 | 3 |
| Overlap | YGR064W | 4.61 | 2.34 | 4 |
| SPT4 |
| ORF | Gene | ΔUBLoaOOH | ΔBIuntreated | LoaOOH sensitivity |
| BY4743 | Wild-type | 17.5 | 0 | Not sensitive |
| YMR169C | ALD3 | 2.79 | −3.27 | 4 |
| YJL115W | ASF1 | 5.05 | 6.51 | 4 |
| YER177W | BMH1 | 5.88 | 4.19 | 4 |
| YER141W | COX15 | 0.38 | 2.11 | 6 |
| YGR036C | CWH8 | 0.31 | 10.13 | 6 |
| YIL065C | FIS1 | 2.29 | −4.2 | 6 |
| YOL051W | GAL11 | −4.44 | 4.24 | 3 |
| YGR163W | GTR2 | 0.09 | 5.65 | 6 |
| YLR192C | HCR1 | 2.33 | 8.3 | 7 |
| YDR174W | HMO1 | 4.36 | 8.33 | 3 |
| YDL115C | IWR1 | 6.56 | 10.96 | 4 |
| YLR244C | MAP1 | 7.72 | 4.46 | 2 |
| YLR320W | MMS22 | 5.51 | −15.91 | 5 |
| YKL009W | MRT4 | 4.15 | 14.49 | 6 |
| YNL099C | OCA1 | 1.18 | 2.36 | 7 |
| YJR073C | OPI3 | 3.02 | 3.93 | 3 |
| YBR035C | PDX3 | 2.1 | 13.3 | 2 |
| YLR148W | PEP3 | 0.82 | 4.29 | 6 |
| YDL065C | PEX19 | 0.32 | −1.25 | 6 |
| YIL107C | PFK26 | 3.43 | −2.1 | 2 |
| YGL025C | PGD1 | −1.74 | −0.13 | 4 |
| YDL006W | PTC1 | 3.36 | 5.89 | 6 |
| YJL121C | RPE1 | 7.8 | −1.36 | 2 |
| YLL002W | RTT109 | −2.46 | −9.45 | 6 |
| YNL032W | SIW14 | 4.33 | 1.63 | 7 |
| YBR289W | SNF5 | 3.36 | 8.1 | 1 |
| YHL025W | SNF6 | 6.49 | 0.93 | 3 |
| YGR104C | SRB5 | −1.85 | 6.92 | 4 |
| YPL057C | SUR1 | 2.66 | 9.44 | 6 |
| YJL176C | SWI3 | 1.33 | 10.63 | 1 |
| YLR182W | SWI6 | 3.08 | 13.75 | 7 |
| YBR069C | TAT1 | 1.52 | 21.51 | 2 |
| YLR237W | THI7 | 2.33 | 5.97 | 7 |
| YCR053W | THR4 | 6.09 | −2.06 | 3 |
| YPR163C | TIF3 | 4.1 | 9.91 | 6 |
| YPR074C | TKL1 | 6.78 | 5.9 | 1 |
| YOR332W | VMA4 | 4.63 | 4.06 | 4 |
| YML007W | YAP1 | 4.11 | 7.3 | 3 |
| YBR285W | YBR285W | 2.33 | 1.63 | 7 |
| YCR061W | YCR061W | 2 | 2.3 | 7 |
| YCR095C | YCR095C | 2 | 3.63 | 7 |
| YDL173W | YDL173W | 5.34 | 1.18 | 3 |
| YIL077C | YIL077C | −6.08 | −0.8 | 6 |
| YLR456W | YLR456W | 4.12 | 1.95 | 4 |
| YNL080C | YNL080C | 1.38 | −3.54 | 5 |
| YOR135C | YOR135C | 6.58 | 1.12 | 3 |
| Overlap | YGR064W | 4.61 | 2.34 | 4 |
| SPT4 |
Data are given as sensitivity to the change in percent budding as a result of treatment with 0.01 mM LoaOOH for one hour (ΔUBLoaOOH). The difference in percentage buds of the untreated wild-type (ΔBIuntreated) is also given, together with the sensitivity of the strains to LoaOOH on a scale 1 (most sensitive) to 7 (least sensitive).
Cellular functions involved in the cell-cycle response to LoaOOH
For each gene identified from the 47 deletant strains, the gene descriptions, over-represented gene ontologies (GO terms) (Dwight et al., 2002) and functional categories (Robinson et al., 2002) were obtained to identify the cellular processes that may be involved in the cell-cycle delay. These are listed in supplementary Table S1a. These included PDX3 and THR4 that encode enzymes involved in vitamin B6 metabolism: how this relates to cell-cycle delay remains speculative. PFK26, RPE1 and TKL1 encode key enzymes involved in glucose metabolism. The pfk26 deletant lacks 6-phosphofructose-2-kinase, which is critical in maintaining the rate of glycolysis. In addition, Rpe1p and Tkl1p catalyse consecutive steps in the conversion of ribulose 5-phosphate into glyceraldehyde 3-phosphate, which are involved in a reversible shuttle of metabolites between the pentose phosphate pathway (PPP) and glycolysis. NADPH produced by the PPP is required for reduction of oxidized glutathione (GSSG) and oxidized thioredoxin to confer protection against damage caused by ROS (Jamieson, 1998). Deletion of TKL1 confers cross-sensitivity to all the major oxidative stresses (Sundstrom et al., 1993) and the PPP has been shown to play protective roles during oxidative stress (Kletzien et al., 1994; Slekar et al., 1996).
It has been noted previously that the putative protein tyrosine phosphatase, Oca1p, is involved in cell-cycle delay in response to LoaOOH (Alic et al., 2001). It has been proposed recently that Oca1p, Ycr095p, Siw14p and two additional proteins Ynl056p and Yhl029p constitute a physical interaction network required for protection against superoxide stress and that these proteins be named Oca1-5p, respectively (Warringer et al., 2005; Ericson, 2006). The occurrence of three mutants of the putative Oca complex in these data indicate that the complex plays a significant role in the cell-cycle delay. Interestingly, Warringer et al., using Synthetic Genetic Array (SGA) analysis, noted strong genetic interactions between members of the Oca complex and Tkl1p, indicating that the Oca complex has an important role in carbohydrate utilization and production of NADH/NADPH, highlighting the potential role of NADPH generation in some aspect of cell-cycle sensing of oxidative damage. It is also interesting that the Oca complex exhibits a strong genetic interaction with Ald6p (Warringer et al., 2005; Ericson, 2006). While the ald6 deletant was not detected as being sensitive to LoaOOH and therefore was not tested, the ald3 mutant was sensitive to LoaOOH and exhibited an altered cell-cycle response to LoaOOH. ALD3 encodes a stress-induced NAD+-dependent aldehyde dehydrogenase that is homologous to Ald6p. Ald3p may play a role in the detoxification of cytotoxic decomposition products of LoaOOH, such as malondialdehyde and 4-hydroxy-2-nonenal (Gutteridge, 1995; Oberschall et al., 2000).
Also prominent in the cellular functions required for cell-cycle delay were eight transcription factors. These included components of Kornberg's mediator (SRB) complex (encoded by PGD1, SRB5 and GAL11) and the SWI/SNF transcription activator complex (SNF5 and SNF6) (supplementary Table S1b). These transcription complexes are a part of the general transcription machinery required for basal and responsive gene expression. The product of SPT4 participates in transcription elongation.
Two specific transcription factors, encoded by SWI6 and YAP1, were also shown to be involved. Of these, the participation of YAP1 is not surprising, given the central role played by Yap1p in the transcriptional response to oxidants and xenobiotics (Coleman et al., 1999). The role of SWI6 is very interesting given the function of Swi6p in cell-cycle progression from the G1 to the S phase, and this aspect is discussed in more detail below. These data indicate that transcription plays an important role in the cell-cycle response to oxidants, in keeping with previous studies that have shown that cell-cycle delay is regulated at the level of transcription (Sidorova & Breeden, 1997).
Interactions between genes involved in the cell-cycle response to LoaOOH
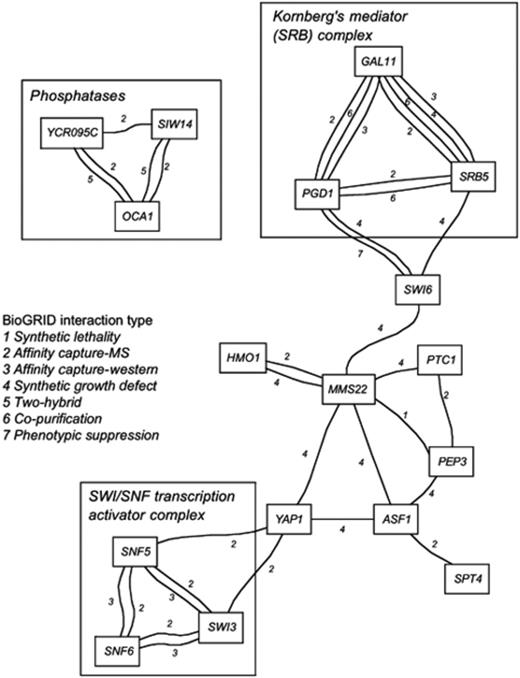
The connections between the functions of many of the 47 genes listed in Table 1 involved in oxidant-induced cell-cycle delay are not obvious from direct inspection of their individual functions. In order to try to identify components of this response that share a common mechanism, all direct known interactions between the products of the genes with reference to the BioGRID database were identified (Stark et al., 2006). The interactions, including protein–protein as well as synthetic lethality and genetic interactions, are represented in Fig. 4. These interactions included three of the over-represented functions from gene ontology data: the SWI/SNF transcription activator complex (Snf5p, Snf6p and Swi3p); Kornberg's mediator (SRB) complex (Gal11p, Pgd1p and Srb5p); and phosphatases (Oca1p, Ycr095p and Siw14p). Of these, the two transcription factor complexes (Kornberg's mediator and SWI/SNF activator complexes) are linked via a set of direct interactions. Yap1p directly interacts with two components of the SWI/SNF transcription activator complex. Yap1p also interacts with other components of the Mediator complex (Med2p, Rox3p and Srb6p) which in turn interact with Gal11p, Pgd1p and Srb5p identified from the data for nonresponsive mutants. These interaction data indicate that Yap1p, which is known to play a role in regulating oxidative stress response genes, acts in conjunction with the mediator and SWI/SNF transcription activator complexes to affect cell-cycle progression. Swi6p also interacts directly with Pgd1p and Srb5 of the Mediator complex.
An interaction network of deletants (nodes) that exhibit the cell-cycle delay phenotype and interaction data derived from the BioGRID database (links). Each interaction type is indicated. Three sets of genes have been boxed and labelled according to their common GO term.
These interaction data raise the possibility of an interaction between Swi6p and Yap1p, linking both oxidant-responsive and cell-cycle responsive gene regulation. Further support for an indirect association between Swi6p and Yap1p is indicated by their common synthetic growth defect interaction with Mms22p, a protein involved in resistance to ionizing radiation (Bennett et al., 2001). Mms22p is a key protein in this interaction network (Fig. 4) because it is the most highly connected member, exhibiting additional synthetic lethality or growth defect interactions with the chromatin-associated high mobility group protein Hmo1p, the type 2C protein phosphatase Ptc1p, the vacuolar peripheral membrane protein Pep3p and the nucleosome-assembly factor Asf1p. Because synthetic genetic analyses often indicate the existence of parallel compensatory pathways, these data may indicate that the cell-cycle response to oxidants can in part be compensated by the Mms22p repair pathway that may resolve replication blocks (Longhese et al., 2003) or by up-regulation of the high osmolarity glycerol (HOG) signalling pathway, which is constitutively active in a strain lacking Ptc1p.
Regulation of cell-cycle progression by the Swi6p transcription factor
The swi6 deletant was sensitive to LoaOOH (Fig. 5a) and it also showed very little change in bud index on treatment with LoaOOH (Table 1). This inability of the swi6 deletant to arrest in the G1/S phase upon treatment with oxidant is particularly interesting, given the well-characterized role of Swi6p as a cell-cycle transcription factor. Swi6p complexes with Swi4p to form the SBF complex, and with Mbp1 to form MBF, either of which can activate cyclin genes required for cell-cycle progression and cell growth (Iyer et al., 2001). In addition, Swi6p interacts with the chromatin-remodelling factors SWI/SNF and Mediator complex to commit the cell to enter the S phase (Li et al., 2005).
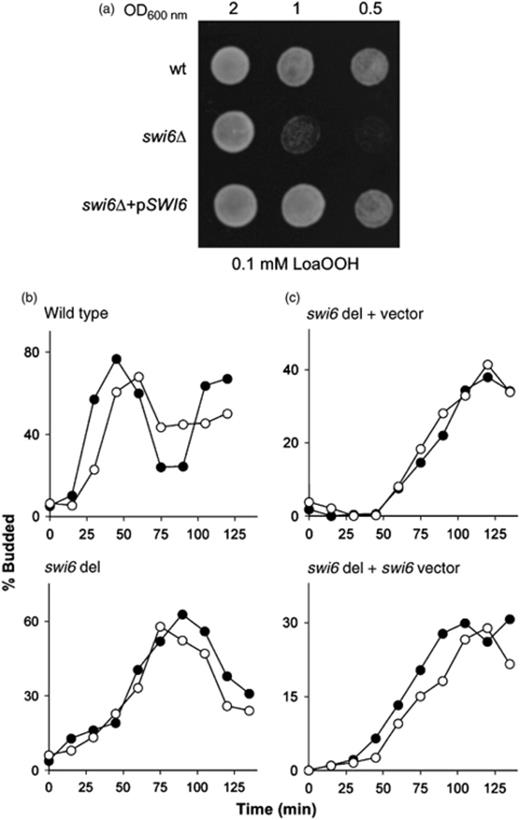
The LoaOOH sensitivity of swi6Δ and its inability to arrest upon treatment is recovered by transforming a single copy of SWI6 on a plasmid. (a) LoaOOH sensitivity of wild-type, swi6Δ and swi6Δ carrying pSWI6 were tested on plates containing 0.1 mM LoaOOH. Stationary phase cell cultures were used for the spot test and photographed after a 48-h incubation. (b) Cell-cycle progression of α-factor synchronized wild-type (top) and swi6 deletant (bottom) strains in response to 0.02 mM LoaOOH. Cell were arrested with α-factor and then washed and treated with LoaOOH. Cells were resuspended in fresh media and aliquots were taken every 15 min to determine the percentage of budded cells. Progression through the cell cycle was determined via the proportion of budded cells viewed under the microscope. Closed circles (●) represent untreated cells and open circles (○) represent LoaOOH-treated cells. (c) The cell-cycle progression of SWI6 deletion transformed with a control centromeric plasmid (top) and the plasmid containing the cloned SWI6-coding sequence and its upstream and downstream regulatory elements (bottom). Representative graphs are shown.
In order to investigate the possible role of Swi6p in the oxidant regulation of cell-cycle progression, the authors characterized in greater detail the cell-cycle phenotype of the wild-type and swi6Δ strains following LoaOOH treatment. Cells were synchronized with α-factor and subjected to treatment with 0.02 mM LoaOOH. The concentration of 0.02 mM was chosen for this experiment after optimization because the conditions used to treat the cells and genetics or background differed from the previous analysis. As shown in Fig. 5b, LoaOOH-treated wild-type cells again exhibited a cell-cycle delay of 15–20 min relative to the wild-type strain. Interestingly, the swi6 deletant showed a delayed entry into the S-phase for 15–20 min after its release from α-factor in the absence of an oxidant. When treated with an oxidant, it did not show any further cell-cycle delay as observed for the wild type. This experiment was repeated before performing the microarray experiment with similar results. To ensure that this loss of cell-cycle delay phenotype was a result of SWI6 deletion, the SWI6-coding sequence and its upstream and downstream regulatory elements on a centromeric plasmid were cloned and transformed into the swi6 deletant. The resulting strain was found to arrest in the G1 phase when treated with LoaOOH (shown in Fig. 5c) in the same way as the wild type. While it is possible that LoaOOH in the swi6 deletion may have slowed release from the α-factor cell-cycle arrest, it is unlikely because the initial screen that identified the swi6 deletant was performed in asynchronous cells (in the absence of α-factor). In addition, while the swi6 deletion is slower to release from α-factor, there is still a differential response between the WT and the swi6 deletant that is restored by the expression of the pSWI6, which is fully consistent with the initial screen data. These data show that the alternative Skn7p-dependent mechanism that allows the swi6Δ strain to progress in the cell cycle (Barrera & Ren, 2006) only does so after a delay in G1 and that this alternative system is not sensitive to LoaOOH treatment.
Because Swi6p is involved with the separate partners Swi4p and Mbp1 in the SBF and MBF complexes, respectively, the responses of swi4Δ and mbp1Δ strains to LoaOOH were also analysed to determine whether either of these complexes was solely responsible for the delay process. Like the wild type, both swi4Δ and mbp1Δ strains accumulated unbudded cells having a ΔUBLoaOOH of 15.30 (SD 0.97) and 13.66 (SD 1.62), respectively, and were thus arrested normally in the G1 phase in response to LoaOOH stress. This indicates that either of the SBF or MBF complexes can function in the wild-type response to treatment with the oxidant, or that there is some unique function or partner of Swi6p that has yet to be identified.
Swi6p involvement in the transcriptional response to LoaOOH
From the above data, it is clear that Swi6p plays a fundamental role in the process of LoaOOH-induced cell-cycle delay. Given the role Swi6p plays in cell-cycle regulation via transcriptional regulation of G1/S phase-specific genes (Iyer et al., 2001), the authors examined the extent to which SWI6 contributed to transcriptional responses to LoaOOH by microarray analyses of the transcripts in the wild-type and swi6 deletant strains before and after LoaOOH treatment.
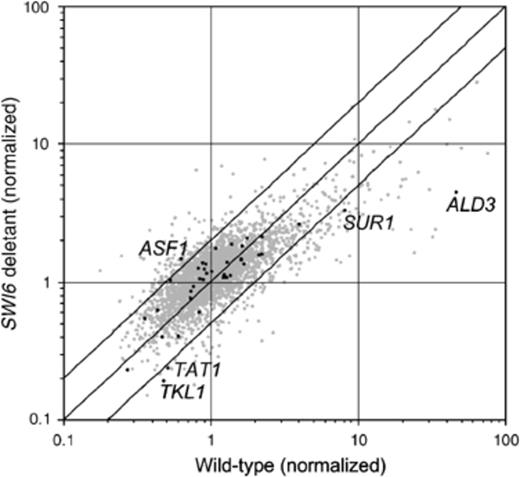
In order to identify genes whose response to LoaOOH treatment depends on Swi6p, the ratio of the expression level of each gene in treated cells compared with untreated cells for the wild-type strain was plotted against the same expression ratio in the swi6 deletant (Fig. 6). In this plot, most genes were clustered along the diagonal, showing that their response to LoaOOH was the same in the wild type and the mutant and was unlikely to be regulated via Swi6p. The absence of SWI6 did, however, have a profound influence on some transcriptional changes induced by LoaOOH. The ratio of treated/untreated transcript levels for 474 genes exhibited at least a twofold difference between the mutant and the wild-type strains; 217 genes were upregulated and 257 were downregulated. The complete set of genes whose expression ratio was perturbed by a greater than twofold in response to LoaOOH in the swi6 deletant relative to the wild type is given in supplementary Table S2. These genes were examined for statistically significant over-represented functional categories using the funspec program (Robinson et al., 2002). The results are given in supplementary Table 3a and 3b. Within these tables, genes whose expression was perturbed by greater than fourfold, and functional categories that are over-represented using only the fourfold data have been highlighted.
Genome-wide transcriptional response to LoaOOH in the swi6 deletant strain compared with the wild type. The genome-wide transcriptional changes resulting from 1 h of treatment with 0.03 mM LoaOOH in the exponentially growing wild type or swi6 deletant cells were determined as described. The normalized ratio (treated/untreated) in the wild type is given on the x-axis, while for the mutant it is shown on the y-axis. The genes whose transcripts were deemed to be present on all slides are shown. The twofold difference boundary of induction in the mutant vs. the wild type is indicated by the outer diagonal lines. The twofold change boundary for the expression in the wild type alone is given by the two outer vertical lines.
These analyses revealed that genes with a reduced response to LoaOOH in the swi6 deletant were enriched for categories representing molecular chaperones, response to stress, large subunit ribosomal proteins and to a lesser extent glucose metabolism (PPP, glycolysis, glucose transporters, positive regulation of glycolysis and gluconeogenesis). However, these genes are not normally associated with regulation by SWI6 and the repression of ribosomal protein genes has been observed previously as part of the (general) environmental stress response (Gasch et al., 2000). The set of genes whose transcripts were upregulated in the absence of SWI6 were enriched for functions including purine ribonucleotide metabolism and to a lesser extent DNA mismatch repair, cell division and the transfer of pentosyl groups. The overall sets of transcripts misregulated in the swi6 deletant relative to the wild-type strain included those whose ratio changed less than twofold in the wild-type strain. This implied that some of the changes observed on deletion of SWI6 occurred to compensate for its absence.
Each of the perturbed gene sets identified from the LoaOOH-treated swi6 deletant was additionally inspected for the occurrence of genes regulated by common transcription factors. This was performed using overrepresentation analysis against gene sets that have common transcription factor-binding interactions at their promoters and correlated gene expression profiles across over 500 microarray experiments (Bar-Joseph et al., 2003). These analyses identify transcription factors involved in regulation, determined by reference to published data describing actual yeast transcription factor binding as opposed to theoretical promoter sequence analyses. This analysis revealed that only five transcriptional regulatory motifs were statistically over-represented (P<0.05) in the perturbed gene-sets. Furthermore, these five sets of potentially coregulated genes were segregated in either the up- or downregulated microarray gene sets as shown in Table 2. Additionally, each of the up- and downregulated gene set from the LoaOOH-treated swi6 deletant was searched for genes that were representative of biochemical pathways (Kanehisa et al., 2004). These analyses revealed that the downregulated gene set was over-represented with genes from seven KEGG pathways (P<0.01) and only a single gene of these entire pathways occurred in the inversely upregulated gene set.
Over-represented genes from various functional categories are highly segregated according to their expression profile in the microarray data from the swi6 deletant treated with LoaOOH
| Expression profile | |||
| Regulatory motif (bound transcription factors) | Down | Up | Over-represented genes in Gene Module |
| BAS1 | 0 | 8 | HIS4 “ADE5,7”MTD1 SHM2ADE13 ADE17 ADE4 ADE12 |
| MBP1, SWI6 | 0 | 3 | MSH6 GIN4 TOF1 |
| IME4, SWI5 | 2 | 0 | HBT1 YHR138C |
| HSF1 | 10 | 1 | HSP26 HSP42SSA4 BTN2YLL023C HSP104 CPR6 YLR327C SIS1 STI1 |
| MSN4 | 5 | 1 | HSP26SSA4 BTN2 YHR087WHSP104 |
| KEGG Pathway | Down | Up | Over-represented genes in KEGG Biochemical Pathway |
| Glycolysis/gluconeogenesis | 10 | 0 | CDC19 GLK1 PGK1 HXK1 TDH3 ENO1 ENO2 TDH2PDC5PGM2 |
| Ribosome | 14 | 1 | RPL4ARPL4B RPS13 SMC2 RPS4B RPL2B RPP0 RPL18B RPP2A RPL18A RPL3 RPL5 RPL7B RPL1A |
| Riboflavin metabolism | 4 | 0 | PHO3 FAD1 RHR2 RIB4 |
| Pentose phosphate cycle | 5 | 0 | YGR043C SOL4 GND1 PGM2 TKL1 |
| Propanoate metabolism | 3 | 0 | LSC1 ALD4 ALD6 |
| MAPK signaling (high osmolarity) | 3 | 0 | CTT1STE20 MSN4 |
| Purine metabolism | 4 | 8 | CDC2 ADK2 “ADE5,7” MET14 ADE13 ADE17 ADE4 ADE12 |
| One carbon pool by folate | 0 | 3 | MTD1 SHM2ADE17 |
| Cell cycle | 3 | 8 | MBP1 CLB3 DBF4 GIN4 MCM6 CDC5 PHO80 DBF20 |
| Expression profile | |||
| Regulatory motif (bound transcription factors) | Down | Up | Over-represented genes in Gene Module |
| BAS1 | 0 | 8 | HIS4 “ADE5,7”MTD1 SHM2ADE13 ADE17 ADE4 ADE12 |
| MBP1, SWI6 | 0 | 3 | MSH6 GIN4 TOF1 |
| IME4, SWI5 | 2 | 0 | HBT1 YHR138C |
| HSF1 | 10 | 1 | HSP26 HSP42SSA4 BTN2YLL023C HSP104 CPR6 YLR327C SIS1 STI1 |
| MSN4 | 5 | 1 | HSP26SSA4 BTN2 YHR087WHSP104 |
| KEGG Pathway | Down | Up | Over-represented genes in KEGG Biochemical Pathway |
| Glycolysis/gluconeogenesis | 10 | 0 | CDC19 GLK1 PGK1 HXK1 TDH3 ENO1 ENO2 TDH2PDC5PGM2 |
| Ribosome | 14 | 1 | RPL4ARPL4B RPS13 SMC2 RPS4B RPL2B RPP0 RPL18B RPP2A RPL18A RPL3 RPL5 RPL7B RPL1A |
| Riboflavin metabolism | 4 | 0 | PHO3 FAD1 RHR2 RIB4 |
| Pentose phosphate cycle | 5 | 0 | YGR043C SOL4 GND1 PGM2 TKL1 |
| Propanoate metabolism | 3 | 0 | LSC1 ALD4 ALD6 |
| MAPK signaling (high osmolarity) | 3 | 0 | CTT1STE20 MSN4 |
| Purine metabolism | 4 | 8 | CDC2 ADK2 “ADE5,7” MET14 ADE13 ADE17 ADE4 ADE12 |
| One carbon pool by folate | 0 | 3 | MTD1 SHM2ADE17 |
| Cell cycle | 3 | 8 | MBP1 CLB3 DBF4 GIN4 MCM6 CDC5 PHO80 DBF20 |
Genes whose expression differed by greater than fourfold are highlighted as bold underlined text.
Over-represented genes from various functional categories are highly segregated according to their expression profile in the microarray data from the swi6 deletant treated with LoaOOH
| Expression profile | |||
| Regulatory motif (bound transcription factors) | Down | Up | Over-represented genes in Gene Module |
| BAS1 | 0 | 8 | HIS4 “ADE5,7”MTD1 SHM2ADE13 ADE17 ADE4 ADE12 |
| MBP1, SWI6 | 0 | 3 | MSH6 GIN4 TOF1 |
| IME4, SWI5 | 2 | 0 | HBT1 YHR138C |
| HSF1 | 10 | 1 | HSP26 HSP42SSA4 BTN2YLL023C HSP104 CPR6 YLR327C SIS1 STI1 |
| MSN4 | 5 | 1 | HSP26SSA4 BTN2 YHR087WHSP104 |
| KEGG Pathway | Down | Up | Over-represented genes in KEGG Biochemical Pathway |
| Glycolysis/gluconeogenesis | 10 | 0 | CDC19 GLK1 PGK1 HXK1 TDH3 ENO1 ENO2 TDH2PDC5PGM2 |
| Ribosome | 14 | 1 | RPL4ARPL4B RPS13 SMC2 RPS4B RPL2B RPP0 RPL18B RPP2A RPL18A RPL3 RPL5 RPL7B RPL1A |
| Riboflavin metabolism | 4 | 0 | PHO3 FAD1 RHR2 RIB4 |
| Pentose phosphate cycle | 5 | 0 | YGR043C SOL4 GND1 PGM2 TKL1 |
| Propanoate metabolism | 3 | 0 | LSC1 ALD4 ALD6 |
| MAPK signaling (high osmolarity) | 3 | 0 | CTT1STE20 MSN4 |
| Purine metabolism | 4 | 8 | CDC2 ADK2 “ADE5,7” MET14 ADE13 ADE17 ADE4 ADE12 |
| One carbon pool by folate | 0 | 3 | MTD1 SHM2ADE17 |
| Cell cycle | 3 | 8 | MBP1 CLB3 DBF4 GIN4 MCM6 CDC5 PHO80 DBF20 |
| Expression profile | |||
| Regulatory motif (bound transcription factors) | Down | Up | Over-represented genes in Gene Module |
| BAS1 | 0 | 8 | HIS4 “ADE5,7”MTD1 SHM2ADE13 ADE17 ADE4 ADE12 |
| MBP1, SWI6 | 0 | 3 | MSH6 GIN4 TOF1 |
| IME4, SWI5 | 2 | 0 | HBT1 YHR138C |
| HSF1 | 10 | 1 | HSP26 HSP42SSA4 BTN2YLL023C HSP104 CPR6 YLR327C SIS1 STI1 |
| MSN4 | 5 | 1 | HSP26SSA4 BTN2 YHR087WHSP104 |
| KEGG Pathway | Down | Up | Over-represented genes in KEGG Biochemical Pathway |
| Glycolysis/gluconeogenesis | 10 | 0 | CDC19 GLK1 PGK1 HXK1 TDH3 ENO1 ENO2 TDH2PDC5PGM2 |
| Ribosome | 14 | 1 | RPL4ARPL4B RPS13 SMC2 RPS4B RPL2B RPP0 RPL18B RPP2A RPL18A RPL3 RPL5 RPL7B RPL1A |
| Riboflavin metabolism | 4 | 0 | PHO3 FAD1 RHR2 RIB4 |
| Pentose phosphate cycle | 5 | 0 | YGR043C SOL4 GND1 PGM2 TKL1 |
| Propanoate metabolism | 3 | 0 | LSC1 ALD4 ALD6 |
| MAPK signaling (high osmolarity) | 3 | 0 | CTT1STE20 MSN4 |
| Purine metabolism | 4 | 8 | CDC2 ADK2 “ADE5,7” MET14 ADE13 ADE17 ADE4 ADE12 |
| One carbon pool by folate | 0 | 3 | MTD1 SHM2ADE17 |
| Cell cycle | 3 | 8 | MBP1 CLB3 DBF4 GIN4 MCM6 CDC5 PHO80 DBF20 |
Genes whose expression differed by greater than fourfold are highlighted as bold underlined text.
The above analysis revealed that eight genes regulated by Bas1p were induced in the swi6 deletant relative to the wild type. Bas1p is a transcription factor involved in the regulation of enzymes of histidine, purine, and pyrimidine biosynthetic pathways and one-carbon metabolism. Of these eight genes (see Table 2), SHM2 and MTD1 exhibit greater than fourfold expression whereas the others exhibit a lesser twofold expression. This relative expression profile is consistent with previous studies of the Bas1p mediated transcriptional regulon (Gelling et al., 2004; Subramanian et al., 2005). These genes are important in the generation of purine nucleotides and may reflect an increased demand for these precursors for DNA repair when progression in the cell-cycle mediated by Swi6p is inhibited. As may be expected, the KEGG metabolic pathways of purine metabolism and one carbon metabolism are also over-represented in this gene set. These data indicate that Swi6p has a direct or an indirect negative regulatory effect on Bas1p. In addition, three genes, MSH6, GIN4 and TOF1, that have been shown to bind MBP1 and SWI6 (Bar-Joseph et al., 2003) are upregulated in the swi6 deletant (as shown in Table 2). This is surprising because SWI6 is an activator of these genes and therefore in its absence these genes should be repressed relative to the wild type. Possibly some compensatory effect of the gene deletion has led to this occurrence. It is also interesting to note that components of the MAPK signalling pathway were down-regulated in the swi6 deletant, indicating a possible involvement of this pathway in the cell-cycle response to oxidant treatment.
A common paradigm of cellular responses is that relevant transcription factors generally function downstream of a signalling pathway or cascade. If this is true for the Swi6p transcription factor-regulated component of the LoaOOH response, then the question arises as to which signalling pathway is involved.
Components of the signalling pathway are poorly represented in the responsive gene-sets
A comparison of the LoaOOH-responsive genes of the wild-type strain (Alic et al., 2004) and the swi6 deletant microarray experiment was performed. There is a high degree of overlap (77 genes) between the upregulated genes of the wild-type strain and those downregulated in the swi6 deletant. These genes are of interest because they represent those activated in the wild type in response to LoaOOH that are conversely downregulated in the swi6 deletant. It is interesting to note that seven out of eight heat shock proteins from the wild-type upregulated set and three out of four proteins involved in glucose transport from the swi6 deletant downregulated set were included in these common 77 genes (Table 3). In addition, there were two instances whereby genes known to be bound by a specific transcription factor (Harbison et al., 2004) occurred almost exclusively within the 77 intersecting genes, for instance 4/5 genes bound by Msn4p and 3/3 genes bound by Stb4p, with respect to the swi6 deletant downregulated gene set, were present in the intersection.
A comparison of the distribution of molecular function and transcription factor binding of the genes down-regulated following exposure to LoaOOH in the swi6 deletant and upregulated in the wild type and the genes common to both sets (the intersection)
| GO term: molecular function | P-value | Intersecting genes | Wild-type upregulated | swi6 downregulated | Genes of intersect |
| Heat shock protein | 8.33E-10 | 7 | 8 | 12 | SSA3 HSP26HSP30HSP78SSA4HSP104HSP82 |
| Glucose transporter | 1.03E-03 | 3 | 5 | 4 | HXT7 HXT6 HXT2 |
| Binding Data | |||||
| MSN4 | 3.32E-03 | 4 | 8 | 5 | HSP26SSA4YHR087WHSP104 |
| STB4 | 5.96E-03 | 3 | 3 | 3 | ZTA1YLL056CGTT2 |
| SUT1 | 3.67E-02 | 3 | 9 | 9 | HXT6 CWP1 HXT2 |
| SNT2 | 4.01E-02 | 2 | 2 | 3 | SSA3 YHR138C |
| YAP1 | 4.18E-02 | 3 | 13 | 5 | SNQ2 OYE2 GTT2 |
| GO term: molecular function | P-value | Intersecting genes | Wild-type upregulated | swi6 downregulated | Genes of intersect |
| Heat shock protein | 8.33E-10 | 7 | 8 | 12 | SSA3 HSP26HSP30HSP78SSA4HSP104HSP82 |
| Glucose transporter | 1.03E-03 | 3 | 5 | 4 | HXT7 HXT6 HXT2 |
| Binding Data | |||||
| MSN4 | 3.32E-03 | 4 | 8 | 5 | HSP26SSA4YHR087WHSP104 |
| STB4 | 5.96E-03 | 3 | 3 | 3 | ZTA1YLL056CGTT2 |
| SUT1 | 3.67E-02 | 3 | 9 | 9 | HXT6 CWP1 HXT2 |
| SNT2 | 4.01E-02 | 2 | 2 | 3 | SSA3 YHR138C |
| YAP1 | 4.18E-02 | 3 | 13 | 5 | SNQ2 OYE2 GTT2 |
The P-value is quoted for the genes of the intersection.
Genes whose expression differed by greater than fourfold are highlighted as underlined text.
A comparison of the distribution of molecular function and transcription factor binding of the genes down-regulated following exposure to LoaOOH in the swi6 deletant and upregulated in the wild type and the genes common to both sets (the intersection)
| GO term: molecular function | P-value | Intersecting genes | Wild-type upregulated | swi6 downregulated | Genes of intersect |
| Heat shock protein | 8.33E-10 | 7 | 8 | 12 | SSA3 HSP26HSP30HSP78SSA4HSP104HSP82 |
| Glucose transporter | 1.03E-03 | 3 | 5 | 4 | HXT7 HXT6 HXT2 |
| Binding Data | |||||
| MSN4 | 3.32E-03 | 4 | 8 | 5 | HSP26SSA4YHR087WHSP104 |
| STB4 | 5.96E-03 | 3 | 3 | 3 | ZTA1YLL056CGTT2 |
| SUT1 | 3.67E-02 | 3 | 9 | 9 | HXT6 CWP1 HXT2 |
| SNT2 | 4.01E-02 | 2 | 2 | 3 | SSA3 YHR138C |
| YAP1 | 4.18E-02 | 3 | 13 | 5 | SNQ2 OYE2 GTT2 |
| GO term: molecular function | P-value | Intersecting genes | Wild-type upregulated | swi6 downregulated | Genes of intersect |
| Heat shock protein | 8.33E-10 | 7 | 8 | 12 | SSA3 HSP26HSP30HSP78SSA4HSP104HSP82 |
| Glucose transporter | 1.03E-03 | 3 | 5 | 4 | HXT7 HXT6 HXT2 |
| Binding Data | |||||
| MSN4 | 3.32E-03 | 4 | 8 | 5 | HSP26SSA4YHR087WHSP104 |
| STB4 | 5.96E-03 | 3 | 3 | 3 | ZTA1YLL056CGTT2 |
| SUT1 | 3.67E-02 | 3 | 9 | 9 | HXT6 CWP1 HXT2 |
| SNT2 | 4.01E-02 | 2 | 2 | 3 | SSA3 YHR138C |
| YAP1 | 4.18E-02 | 3 | 13 | 5 | SNQ2 OYE2 GTT2 |
The P-value is quoted for the genes of the intersection.
Genes whose expression differed by greater than fourfold are highlighted as underlined text.
Twenty-five genes were common to those downregulated in the wild-type strain and upregulated in the swi6 deletant. These are genes that exhibit opposing expression profiles in response to LoaOOH and these changes are dependent on the deletion of Swi6p. These genes were poorly enriched for any GO terms; however, further analysis revealed overrepresentation of genes that show stringent binding (P<0.001) of both Swi4p and Swi6p (HO, GIC2, MNN, 1 ERG3 and SVS1) and Rlm1p transcription factors (GIC2 and MNN1) (Harbison et al., 2004). This is interesting because these transcription factors are the downstream effectors of the Mpk1p (Slt2p)-mediated MAPK signalling pathway defined by the KEGG metabolic pathway 04010sce (Kanehisa et al., 2004). These data indicate that the wild-type response to LoaOOH involves the Swi6p-mediated down-regulation of the Mpk1p-mediated MAPK signalling pathway because in the swi6 deletant genes regulated by this pathway are no longer repressed. Furthermore, the intersection of the downregulated genes of the wild-type strain and similarly responsive downregulated genes of the swi6 deletant gave rise to only nine genes; however, two of these genes, PMA1 and GND1, encode proteins that physically interact with Mpk1p. Again, these data indicate a role for Mpk1p in the cellular response to LoaOOH. This is consistent with the finding that Mpk1p is required for the full resistance to LoaOOH, and that the protein is rapidly and transiently activated in response to LoaOOH (Alic et al., 2003). It was, however, shown by these authors that Mpk1p is not directly involved in cell-cycle delay induced by LoaOOH, despite the fact that oxidative stress due to hydrogen peroxide has been shown to activate Mpk1p (Vilella et al., 2005).
Discussion
This study has identified 47 deletant strains that exhibit an inability to delay at Start, in response to assault by LoaOOH. These data implicate that the deleted gene products covering a broad range of cellular functions are required for the G1 checkpoint to operate, whether by a direct or an indirect mechanism. This nonresponsive phenotype is highly prevalent in the subset of deletants that are highly sensitive to LoaOOH (Thorpe et al., 2004), leading to the conclusion that the inability of strains to mount a checkpoint response to low doses is strongly associated with the sensitivity of these strains at higher doses.
The screening data show that components of the Mediator complex are required for the phenotypic response. It is known that the Mediator complex is required for diverse aspects of transcription, such as activation, repression and stimulation of basal transcription. The diversity of these functions has raised the possibility that the Mediator complex contains functionally distinct modules. The existence of such modules was demonstrated previously with the finding of the Rgr1p and Srb4p subcomplexes and the Gal11p module within the Rgr1p subcomplex as an activator binding module (Kang et al., 2001).
The Yap1p transcription factor is crucial for the normal response of cells to a variety of stress conditions including oxidative stress, stress mediated by many drugs and heat shock. Importantly, Yap1p is primarily cytosolic, but after exposure of cells to a number of different oxidizing agents, such as H2O2, diamide and diethylmaleate, the protein rapidly accumulates in the nucleus (Kuge et al., 1997) to increase the expression of a diverse range of oxidant defence genes. The finding that Yap1p may play a crucial role in the phenotypic response to LoaOOH indicates a connection between a major stress-regulated transcription factor and the regulation of the cell cycle. It remains to be investigated which genes are directly controlled through the Yap1p transcription factor in response to LoaOOH in order to understand its precise role in the pathway of the stress-induced cell-cycle modulation.
It is interesting that the swi6 deletant exhibited the no-delay phenotypic response; however, deletion of its canonical binding partners Swi4p and Mbp1p did not. These data indicated that there is a sufficient degree of redundancy between the SBF and MBF complexes for cells to exhibit wild-type characteristics. It would be interesting to determine the phenotypic response of the swi4 mbp1 double mutant to LoaOOH; however, this mutant was not viable in the BY strain background, which is consistent with previous attempts to create this strain (Koch et al., 1993). Alternatively, SWI6 may interact with another factor in response to LoaOOH to activate the cell-cycle delay.
Five genes identified in the cell-cycle screen exhibited perturbed regulation in the swi6 deletion strain after LoaOOH treatment. Of these, ASF1 occurred in the up-regulated set while TAT1, ALD3, SUR1 and TKL1 occurred in the downregulated set, as shown in Fig. 6. Asf1p, which is known to be involved in the induction of apoptosis, may be suppressed by Swi6p because its expression is higher in the swi6 deletant than in the wild type after the LoaOOH treatment. The gene encoding Ald3p, a aldehyde dehydrogenase was expressed up to fourfold higher in the wild type after LoaOOH treatment compared with its expression in the swi6 deletant; these data indicate that Swi6p is required for Ald3p induction in response to LoaOOH. ALD3 is one of five genes known to encode aldehyde dehydrogenases in S. cerevisiae. The expression of ALD2 and ALD3 is dependent on the general stress transcription factors Msn2p and Msn4p but independent of the HOG MAP kinase pathway. Ald3p is induced by a variety of stresses, including osmotic shock, heat shock, glucose exhaustion, oxidative stress and drugs. ALD2 is only induced by osmotic stress and glucose exhaustion (Navarro-Avino et al., 1999).
Because Swi6p has no DNA-binding domain and yet is central to the two G1/S phase transcription complexes, it has been suggested that the Swi6p is a target of regulation. While components of the MAPK-signalling pathway have been identified in these data, there is no further indication that components upstream of Mpk1p are involved and Mpk1p is not required for the cell-cycle delay in response to LoaOOH. One possibility is that Swi6p is itself able to respond directly to oxidative stress without the need for, or additional to, a canonical signal transduction pathway. In addition, while 77 genes were upregulated in the wild-type strain and downregulated in the swi6 deletant when treated with LoaOOH, it is interesting that many other gene expression profiles were not significantly different between the two strains. Hence, the deletion of SWI6 does not account for all the changes observed in the wild type, indicating the existence of other mechanisms regulating cell-cycle arrest induced by oxidative stress.
Acknowledgements
This work was supported by grants from the Australian Research Council and by an Australian Postgraduate Award to N.A.
References
Supplementary material
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1567-1364.2007.00349.x (This link will take you to the article abstract).
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.