-
PDF
- Split View
-
Views
-
Cite
Cite
Nadine Camougrand, Ingrid Kiššová, Gisèle Velours, Stéphen Manon, Uth1p: a yeast mitochondrial protein at the crossroads of stress, degradation and cell death, FEMS Yeast Research, Volume 5, Issue 2, November 2004, Pages 133–140, https://doi.org/10.1016/j.femsyr.2004.05.001
Close - Share Icon Share
Abstract
UTH1 is a yeast aging gene that has been identified on the basis of stress resistance and longer life span of mutants. It was also shown to participate in mitochondrial biogenesis. The absence of Uth1p was found to trigger resistance to autophagy induced by rapamycin. Uth1p is therefore the first mitochondrial protein proven to be required for the autophagic degradation of mitochondria. Since this protein is also involved in yeast cell death induced by heterologous expression of the pro-apoptotic protein Bax, the results are discussed in the light of evidence suggesting a co-regulation of apoptosis and autophagy in mammalian cells.
1 Introduction
A coordinated sequence of cell growth, proliferation and death occurs during the life of a cell. Moreover, growth factors, nutrients and stress signals influence cell physiology. Mitochondria have long been considered to play a straightforward but critical role in the life of the cell, namely to carry out energy-yielding oxidative reactions that create the vast majority of ATP necessary to support all cellular functions.
Among the various theories attempting to explain the aging process, that related to free radicals, first proposed by Harman [1], has received most attention until now. The basis of this theory is that reactive oxygen species (ROS) are produced as a normal by-product of aerobic life and that the accumulation of oxidative damage due to ROS induces the fundamental changes responsible for aging. Approximately 90% of the oxygen consumed within a eukaryote is used in mitochondrial respiration, so that the metabolic rate of a cell is tightly related to mitochondrial function and the amount of mitochondria within an aerobic cell or tissue can be used as an indirect index for metabolic rate. Consequently, ROS production within any given cell is essentially dependent on mitochondrial function and on the level of antioxidant defenses. The asymmetric inheritance of oxidation-damaged proteins that have accumulated with replicative aging may contribute to free-radical defense and the fitness of newborn cells [2]. Numerous investigations have demonstrated a clear mitochondrial basis for aging in filamentous fungi, yeast and other species including humans. Although the molecular mechanisms involved in the control of aging may differ in their details from one system to another, it is clear that mitochondrial oxidative stress plays a major role in life span control in various systems. Consequently, aging is associated with a marked decline in mitochondrial function, characterized by the decrease in oxidative phosphorylation and ATP synthesis, the increase in mtDNA mutations, the appearance of abnormal mitochondrial cristae and a marked rise in free-radical production [3]. Mitochondria undergo frequent fission and fusion events that regulate their morphology, number and function [4]. Mitochondrial fission is required in dividing cells to ensure inheritance of organelles by daughter cells, but it is also important during differentiation and in response to new energy demands. Mitochondrial fusion serves to maintain a tubular mitochondrial network and optimal mitochondrial function. Consequently, a balance between mitochondrial fission and fusion events is required for normal mitochondrial and cellular function. Excessive mitochondrial fission and a lack of fusion result in breakdown of the mitochondrial network, loss of mtDNA, respiratory defects and an increase in ROS production [5]. Emerging evidence suggests that mitochondrial fission might also be an important part of the cell death machinery [6,7]. In all these cases, it would be expected that cells have developed a system to remove damaged mitochondria.
Under natural conditions, yeast often faces environmental changes requiring a modulation of its mitochondrial content [reviewed in 8–10]. For example the shift from a respiratory carbon source to a fermentative carbon source or, more drastically, the shift from aerobic to anaerobic conditions, is accompanied by a decrease of both the amount and the enzymatic equipment of mitochondria. Clearly, the inhibition of mitochondrial biogenesis is likely to play an important role in these changes [11,12] but might not be enough to explain the rapid disappearance of cohorts of mitochondria. One study, based on a null mutant of the mitochondrial protease Yme1p, suggests that some mitochondrial alterations are able to trigger mitochondrial autophagy [13,14]. Moreover, a study reported the presence of mitochondria in autophagosomes following autophagy induction by the TOR-kinases inhibitor rapamycin [15]. But hypotheses about the role of mitochondria in their own degradation remain highly speculative since the molecular support is still lacking [16].
2 The UTH1 gene
The UTH1 gene was isolated in three independent genetic screens. The first one was based on isolation of strains exhibiting increased life span during starvation stress. In yeast, life span is defined by the number of cell divisions undergone by mother cells before they stop dividing. Consequently, in yeast life span is more accurately characterized by the number of progeny that an individual cell produces rather than by chronological age; the mother cell dies after a finite number of divisions. The genetic analysis of yeast aging was initiated by the cloning of genes that are differentially expressed during the life span; such differential expression constitutes a secondary phenotype for a longevity gene [17]. A number of genes that influence longevity in yeast have already been identified.
The screening by Kennedy et al. [18] has led to the identification of four “youth” genes (UTH genes) [18,19]. One of them, SIR4 (UTH2) has several functions including chromatin silencing [20] and repression of genes placed near telomeres [21]. UTH1 was another of the genes involved in life span identified in this study.
The UTH1 gene was also identified by screening a Saccharomyces cerevisiae promoter-probe gene bank for oxidative-stress-responsive genes [22], in the absence of which cells showed altered sensitivities to oxidative damage.
A third genetic screening designed to find yeast genes required for death induced by the heterologous expression of the mammalian pro-apoptotic protein Bax has also allowed to identify the UTH1 gene [23].
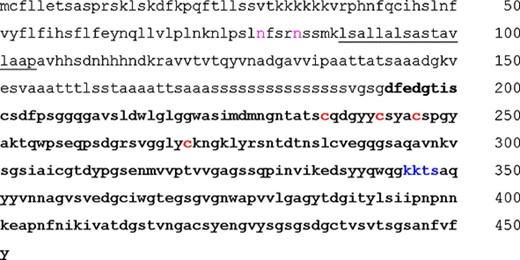
The UTH1 gene belongs to the family of yeast genes termed the “SUN family” (based on the founding members SIM1, UTH1, NCA3, SUN4/SCW3) that has been characterized from different approaches and has been defined on the basis of a common 258-amino-acid C-terminal domain of their gene product (75–85% identity). Actually, a search in databases revealed homologues of the “SUN family” proteins in other fungi such as Candida wickerhamii, C. albicans, Schizosaccharomyces pombe and Neurospora crassa, most of which are thought to be involved in cell wall integrity. The deduced Uth1p protein has a calculated molecular weight of 46.9 kDa, is serine-rich (15% aa) and contains 36.6% non-polar, 25.3% polar, 8% basic and 6.9% acidic amino-acid residues. According to its hydropathy profile [24], the protein may contain a potential transmembrane domain around position 100. Uth1p also contains a potential site of phosphorylation and two potential sites of N-linked glysosylation. One region comprises a potential Fe–S cluster with four putative iron-binding cysteine residues in a Cys-X5-Cys-X3-Cys-X24-Cys array, that is conserved in the SUN family (Fig. 1).
Representation of the primary structure and functional sites in Uth1p. The high homology domain with other “SUN” family members, corresponding to the 258 C-terminal residues is in bold. The potential transmembrane α-helix (residues 88-104) is underlined. Potential N-glycosylation sites (N79 and N83) are in pink. The potential protein-kinase A-dependent phosphorylation site is in blue. The four cysteine residues forming a potential iron/copper binding site are in red.
The four SUN proteins are involved in very different cellular functions [25–28]. Two of these proteins were localized in mitochondria: outer mitochondrial membrane for Uth1p and matrix compartment for Sun4p [29].
Inactivation of UTH1 promotes pleiotropic effects at different cellular levels such as aging, oxidative stress response and mitochondrial biogenesis. UTH1 has been demonstrated to interfere with mitochondrial biogenesis [26]. In cells inactivated for the UTH1 gene, cytochromes aa3, c and b were lowered by 25%, 20% and 15%, respectively, and measurements on whole lysed cells showed that another mitochondrial enzyme, citrate synthase, was also lowered. Electron micrographs revealed no difference in global mitochondria content, and cardiolipid and mtDNA quantifications were equivalent in parental and Δuth1 strains. These results suggest that the protein equipment could be lowered in mitochondria from strains inactivated for UTH1. Despite this decreased content in mitochondrial complexes, strains inactivated for UTH1 were not affected in their doubling time and exhibited a higher growth yield.
3 Response to different stresses
Depletion of UTH1 led to modifications in the response to different stresses. A higher tolerance of Δuth1 cells to growth at 37 °C and resistance to a heat stress (incubation of cells for 1 h at 55 °C) compared to the parental strain were observed. A similar resistance of Δuth1 strain was observed under carbon or nitrogen starvation conditions.
Inactivation of UTH1 increased the sensitivity of cells to copper and paraquat (a drug generating superoxide radicals) and their resistance to H2O2. These effects were independent of the carbon source used in the growth medium (glucose or lactate). These results were in accordance with those obtained by Bandara et al. [22]. The opposite phenotypes of the Δuth1 mutant towards paraquat and H2O2 stresses provide support for the existence of differences in the specificity of the stress responses mediated by the two oxidants [30,31]. In aqueous neutral pH solutions, superoxide ion can behave as a reducer of oxidized transition metal complexes of Fe3+ and Cu2+, whereas H2O2 acts as an oxidant. The well-documented Haber–Weiss and Fenton reactions show the reduction of Fe3+ by superoxide ion and reoxidation of Fe2+ by H2O2[32]. The basis of the differential response of the Δuth1 mutant to the two ROS may result from their differential effects on metal ions. This is interesting considering the possibility of Uth1p to be an iron or copper-binding protein due to the presence of a putative Fe–S cluster, with a potential effect on the metal homeostasis in the cell.
If free radicals and oxidative stress are at least partly responsible for lifespan and aging, it follows that antioxidants should prolong life and retard aging. Indeed, some supporting evidence has been observed. Bacteria and yeasts have been used for many years as simple model systems to study the function of antioxidant enzymes. The first report on the yeast chronological life span model system showed that cytoplasmic and mitochondrial superoxide dismutases, but not catalase or metallothionein, are required for the long-term survival of yeast. Sod2p (mitochondrial enzyme) was found to be required under both low and normal oxygen conditions, whereas cytoplasmic Sod1p was mainly required under normal aeration [33]. A good correlation has also been observed between SOD concentration and the maximum life span of species [34]. In Δuth1, neither differences in ROS production nor modifications in SOD and catalase activities were observed compared to the wild-type strain. Consequently, the increase in life span observed in Δuth1 could not be simply explained by a change in the oxidative stress response, and results described above show that Uth1p is not involved in a general oxidative response pathway.
4 Uth1p is required for Bax-induced cell death
Bax-induced cell death in yeast exhibits several characteristics that depend on metabolic orientation and, more precisely, on the degree of mitochondrial differentiation. When grown on a strictly respiratory carbon source, such as lactate, yeast carries well-differentiated mitochondria. Under these conditions, Bax expression induces a rapid release of cytochrome c from mitochondria to cytosol [35,36]. This release probably correlates with the formation of a giant channel similar to the Mitochondrial Apoptosis-induced Channel (MAC) which appears in mammalian apoptotic cells [37,38]. As yeast is a facultative aerobe it can grow, even when mitochondria are impaired for oxidative phosphorylation, on a fermentable carbon source. Under these conditions, Bax expression also leads to cell death, although far less rapidly than under respiratory conditions [39]. Importantly, Bax-induced cell death occurs in a cytochrome c-less strain or under conditions in which cytochrome c is not released, showing that Bax-induced cell death in yeast does not only depend on cytochrome c release [39,40]. Apoptosis-like characteristics have been observed during Bax-induced cell death, namely chromatin fragmentation and phosphatidylserine exposure [41]. Concerning plasma membrane permeability, Bax expression protects the plasma membrane against ethanol-induced permeabilization, thus shifting a “necrotic type” of death towards an “apoptotic type” of death [42]. Similar characteristics have been observed in yeast submitted to Bax-unrelated death inductions, such as cdc48 mutation [43], moderate oxidative stress [44], arl1 mutation [45], acetic acid treatment [46] and senescence [47]. An observation was reported recently showing that the “apoptosis-like” response following H2O2 treatment is under the control of a caspase-like protease [48]. These data support the idea that different treatments are able to trigger in yeast a type of death that differs from mammalian apoptosis, but nonetheless exhibits overlapping characteristics with apoptosis. The molecular mechanisms involved in this form of death remain to be elucidated.
A strain in which the UTH1 gene was deleted exhibited resistance to Bax expression. The absence of Uth1p did neither change the mitochondrial localization of Bax, nor its insertion in the mitochondrial outer membrane, nor its cytochrome c-releasing activity [23]. On the other hand, the absence of Uth1p prevented the appearance of other hallmarks related to Bax expression in yeast, such as oxidation of mitochondrial lipid, production of reactive oxygen species and maintenance of plasma membrane properties after ethanol stress [23]. The main conclusions of these results are that bax expression induces yeast cell death in two ways: one depends on Uth1p and leads to a cytochrome c-independent autophagic cell death (see below) and the other correspond to an “apoptosis-like” cell death involving the formation of MAC, cytochrome c release, possibly oxidative stress, and activation of caspase-like proteases (Fig. 2).
Schematic representation of the effect of mammalian pro-apoptotic protein Bax on yeast mitochondria. On the left, Bax is involved in the formation of MAC, which favors the release of cytochrome c in the cytosol, like in mammalian apoptotic cells [37,38]. The presence of cytochrome c in the cytosol activates, either directly or indirectly by unbalancing the redox potential, the caspase Yca1p, which is responsible for “apoptosis-like” cell death [48]. On the right, Bax activates the autophagic death pathway which involves Uth1p [23], which is normally activated by the TOR-dependent signalling pathway, but distinct from macroautophagy [Kiššová I, Manon S, Camougrand N, unpublished data].
5 Involvement of Uth1p in autophagic degradation of mitochondria
The most interesting aspect in the function of Uth1p is its involvement in the response to rapamycin. The treatment of cells with rapamycin induces autophagy in yeast and mimics the nitrogen starvation response even under nutrient-rich conditions [49]. Macroautophagy in yeast has emerged as a crucial degradation process leading to intracellular reorganization in peculiar conditions such as nitrogen starvation [16]. It involves the non-selective sequestration of large portions of the cytoplasm into double-membrane structures termed autophagosomes, and their delivery to the vacuole for degradation. Although the normal function of macroautophagy is thought to provide amino acids to starved cells, evidence suggests that it could also be required for the elimination of selected organelles, namely mitochondria, during transitions between environmental conditions.
In mammalian cells, autophagic removal of mitochondria has been shown to be triggered following a process of induction/blockade of apoptosis [50,51]. Also, apoptosis and autophagy can be co-regulated by the Akt/PKB-signalling pathway in mammalian cells [52]. Finally, molecular data suggest that autophagy-specific proteins might also be involved in the regulation of apoptosis [53]. Since mitochondrial events are now regarded as a crucial step in apoptosis, the idea indicates that damaged mitochondria (following apoptotic induction) could control their own degradation by an autophagic process [54]. This is supported by the observations that overexpression of anti-apoptotic proteins, such as Bcl-2, is able to block autophagy [55] and that down-regulation of Bcl-2 induces autophagy [56]. Also, the opening of mitochondrial Permeability Transition Pore (mPTP), often considered as a crucial event regulating the initial steps of apoptosis, induces the colocalization of mitochondrial and lysosomal markers, which is typical of an autophagic process in mammalian cells [57]. In yeast, a study reported the presence of mitochondria in autophagosomes following autophagy-induction by the TOR-kinases inhibitor rapamycin [15].
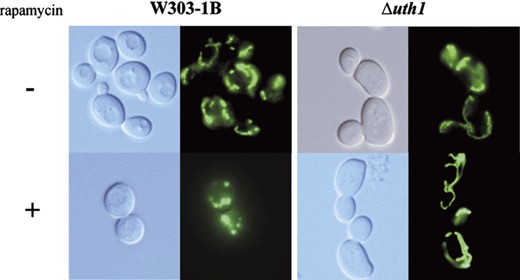
In yeast grown under strict respiratory conditions, a rapid degradation of mitochondria by rapamycin could be visualized. Within 2 h of rapamycin treatment, mitochondrial proteins from different sub-compartments are degraded in a way which depends on autophagosomes formation and vacuolar degradation. This degradation of mitochondrial proteins correlates with the disappearance of the mitochondrial network, but this event did not appear in the Δuth1 mutant (Fig. 3). Also, degradation of the mitochondrial proteins porin and cytochrome oxidase subunit 2 (cox2) was followed in comparison to the cytosolic protein phosphoglycerate kinase (PGK): both mitochondrial proteins were found to be more rapidly degraded in wild-type cells as compared to PGK, whereas in the Δuth1 mutant, the amounts of porin and cox2 remained unchanged during at least 4 h of rapamycin treatment (data not shown). These data are strongly consistent with the hypothesis that the autophagic degradation of mitochondria is impaired in the mutant strain.
Resistance of the mitochondrial network to autophagic degradation in the absence of Uth1p. The wild-type and Δuth1 strains expressing a GFP targeted to the mitochondrial matrix were treated with rapamycin for 2 h. The degradation of the mitochondrial network in the wild-type strain did not appear in the mutant strain.
6 Conclusion
UTH1 is the first known yeast gene providing a link between stress response, aging and mitochondria [58]. It has been shown that it is involved in the regulation of mitochondrial biogenesis. UTH1 overexpression is lethal for cells while its deletion increases their life span. This expanded life span could be correlated with the decrease in mitochondrial biogenesis observed in Δuth1 mutants, leading to a higher efficiency of the respiratory chain. Moreover, the dual involvement of Uth1p in the response to Bax expression and rapamycin treatment suggests that this mitochondrial protein might be at the crossroads of different signals converging to mitochondria and leading to death (Fig. 2).
Since normal cell growth and development can only be based on a homeostatic balance between synthesis and degradation, protein degradation and organelle turnover are obviously as important as protein synthesis and organelle biogenesis. For turnover of cellular components, eukaryotic cells are equipped with several degradation systems, one of them being autophagy. The term autophagy covers distinct, but related, dynamic phenomena that take place in all eukaryotic cells. The best characterized, and the less specific, is the macroautophagy of large parts of the cytoplasm, including the organelles [16]. But more selective phenomena have also been evidenced, such as microautophagy of the nucleus via the physical contact between Nvj1p on the nuclear envelope and Vac8p on the vacuolar membrane [59]. This process does not involve the components of macroautophagy-signalling but is still under the control of the TOR-kinases pathway.
Induction of autophagy has been observed in numerous diseases including neurodegenerative affections [60], myopathy [30], and invasion by microbial pathogens [61]. It is clear that mitochondria are crucial for different processes such as energy production, stress response, apoptosis and aging. Since mitochondrial damages are expected to be critical for the cell, an autophagic process would play an important role to cure cells from injured mitochondria and to regulate their turnover. It is noteworthy that recent data suggest that the “apoptosis-like” pathway is involved in the elimination of injured yeast cells from a senescent population [62]. Within this view, the role of Uth1p-dependent mitochondrial autophagy would take place one step before, when cells can still survive provided that injured mitochondria are removed.
The finding that Uth1p is a component required for the degradation of mitochondria is now a basis for the identification of other components involved in this process. This might be of general interest, not only in yeast, but also, given the recent evidence for correlations between apoptosis and autophagy, in mammalian cells [52,63].
Acknowledgements
This work was supported by grants from the Centre National de la Recherche Scientifique, the Association pour la Recherche contre le Cancer, the Conseil Régional d'Aquitaine and the Université Victor Ségalen Bordeaux 2, and a post-doctoral fellowship from the Association pour la Recherche contre le Cancer (to I.K.).
References
Author notes
Permanent address: Comenius University, Faculty of Sciences, Bratislava, Slovak Republic.



![Schematic representation of the effect of mammalian pro-apoptotic protein Bax on yeast mitochondria. On the left, Bax is involved in the formation of MAC, which favors the release of cytochrome c in the cytosol, like in mammalian apoptotic cells [37,38]. The presence of cytochrome c in the cytosol activates, either directly or indirectly by unbalancing the redox potential, the caspase Yca1p, which is responsible for “apoptosis-like” cell death [48]. On the right, Bax activates the autophagic death pathway which involves Uth1p [23], which is normally activated by the TOR-dependent signalling pathway, but distinct from macroautophagy [Kiššová I, Manon S, Camougrand N, unpublished data].](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/femsyr/5/2/10.1016_j.femsyr.2004.05.001/1/m_FYR_133_f2.jpeg?Expires=1716640328&Signature=WSsAr9RaIS-LUgbDRE5YioYbqN7IbQkaG9VKFxS-bixQlxhfIz~znbzBqeLUYB3QNG76wxpPNN5B9SZ2XROP5cOpPKjSgO14eqIbjYGvpRBfWDqGSdN5G-2xvPUDhKGwS8nRdvNdmW~h23rwhLiT1VNOOMjrBHgQjHSPItStmVFpswG-SzA6qsJ7~ZMrh8WhvCeefheXF9sL1tHQASgg4SWLdEG7MIRCBsG~Be9lh0n0nQUNK-vF-E2fJyl8It8xa8E8sh-OcGwkCPoYpeffl7EDVjv7L3~gqz-iHivCzmsL5OfYyrUQlrI7SPI8tq8mGpqmV4s~cprMuskaLbxSAg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)